There are at least 15 countries now running genome sequencing projects. The team in Manchester, UK, and Boston, USA, share their SEQaBOO project.
Abstract
SEQaBOO (SEQuencing a Baby for an Optimal Outcome) will transform newborn hearing screening (NBHS) by bringing genomic sequencing into routine clinical care, making it possible to identify novel genetic variants for deafness and paving the way to personalised treatments for babies who are deaf or hard of hearing (DHH).
Background
In the United States, 98.2% of newborns receive universal NBHS and 1.7 newborns per 1000 births are DHH [1]. Permanent deafness affects the way that newborns develop by slowing the development of language and social interactions, subsequently affecting quality of life [1]. Identifying deafness early, through NBHS, and providing early intervention are the most effective treatments for babies who are DHH. The success of NBHS has greatly reduced the age of a DHH diagnosis and early intervention, with the current US recommendation being a diagnosis before three months of age and an intervention before six months of age. Current interventions are, however, generic – hearing aids (HA) and cochlear implants (CI) – and may not be effective, highlighting the need for personalised medicine to treat the specific cause of deafness [2].
“More than 150 genes and 8000 pathogenic variants are associated with deafness, with new genes and variants being identified each year”
Personalised medicine offers individual, customised treatments for patients, considering, wherever possible, their genetic information. It is gaining interest globally with at least 15 countries delivering new genomic sequencing projects, or expanding on previous successes [3]. It was supported in the US by President Obama in 2015 through what has become the ‘All of Us’ initiative, and in the UK through the Genomic Medicine Service, as a result of the 100,000 Genomes Project. Personalised medicine may involve genome sequencing – looking at all of a person’s DNA; exome sequencing – looking at the protein-coding regions (exomes) of DNA only; or gene panel screening – looking at a selection of genes that are known to cause or contribute to a medical condition.
SEQaBOO
SEQaBOO is bringing personalised medicine to routine clinical care for newborns who are DHH, and is running in parallel in the US (Boston) and UK (Manchester). It will produce a profile of coding and non-coding pathogenic variation(s) in DHH newborns, including biological parents, wherever possible, to find out how deafness is inherited. More than 150 genes and 8000 pathogenic variants are associated with deafness, with new genes and variants being identified each year. This high level of genetic variation is not seen in other medical conditions and supports the need for the personalised genetic diagnoses that SEQaBOO will provide, ultimately leading to personalised genetic therapies. Further, the frequency and distribution of the genes and variants that cause deafness differ across and between ethnic populations [4], supporting the need to include genetic heritage in diagnostic and therapeutic options for DHH patients. At this early stage of establishing a genetic diagnosis for deafness, however, families should understand the current limitations of genomic research and be are aware that a genetic diagnosis may not be identified, and/or that a genetic diagnosis may not provide a direct personalised treatment. Many genetic variants are unique to a family and their validation is not yet rigorously established. SEQaBOO is a long-term investment for audiological diagnoses and treatment innovation.
“The development of precise, targeted cell- and gene-based therapies to restore or improve hearing will transform the lives of patients who are DHH”
SEQaBOO Boston
SEQaBOO Boston uses genome sequencing to identify known and novel variants that may contribute to/cause deafness, while also offering an opportunity for families to receive secondary feedback on the American College of Medical Genetics and Genomics disease-causing variants in 73 genes (known as the ACMG secondary findings (SF) v3.0 [5]). The ACMG SF v3.0 are largely associated with inherited cancers and heart conditions, and knowing about these variants in an individual can help them to manage their lifestyle and reduce their risk of associated conditions. Participation in the ACMG SF v3.0 is optional and families may prefer to look only for genetic variations that are associated with deafness.
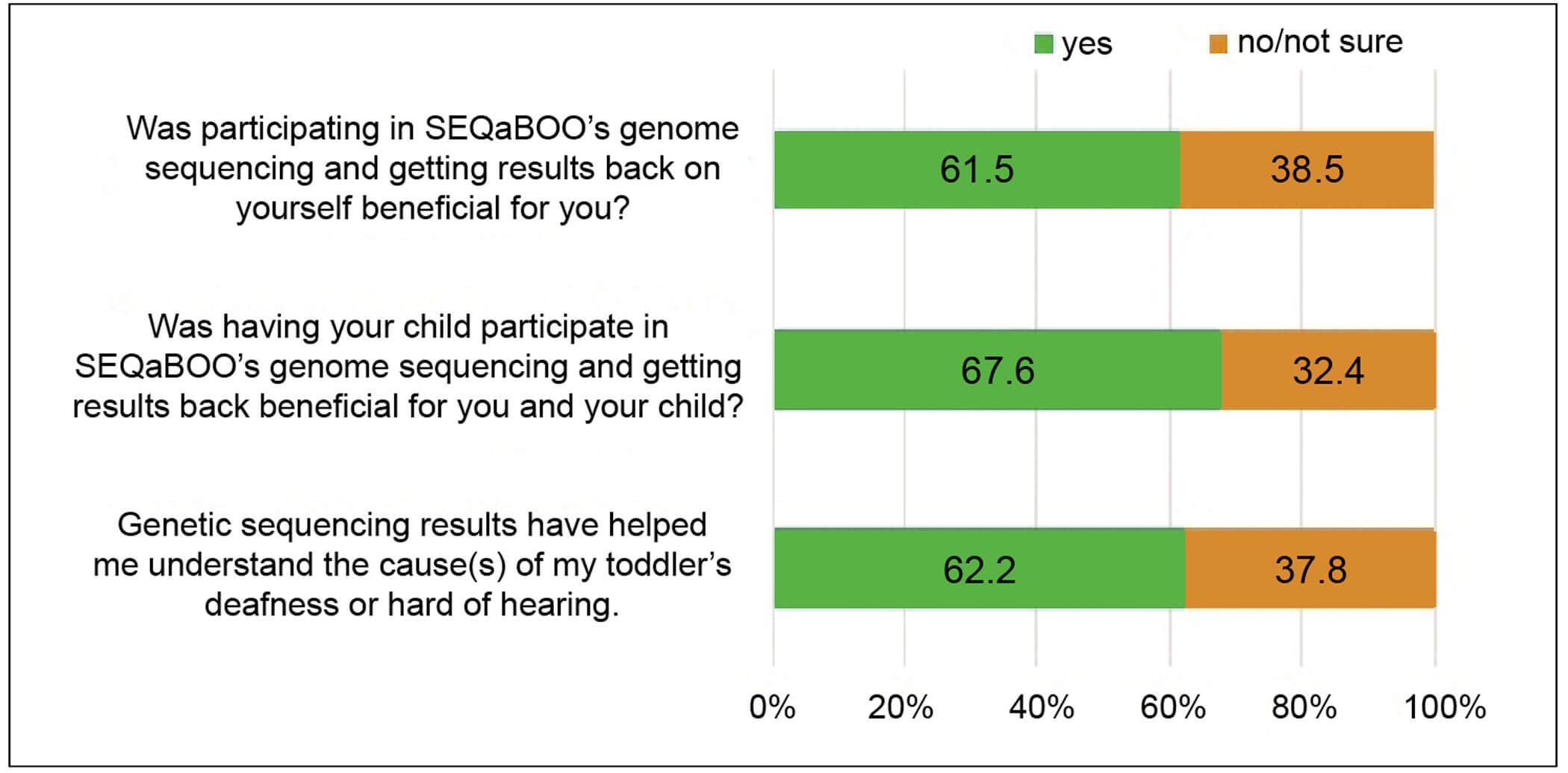
Figure 1. Parental feedback on the benefits of participating in SEQaBOO Boston.
Participants are being recruited from three Harvard Medical School affiliated hospitals: Brigham and Women’s Hospital, Boston Children’s Hospital, and the Massachusetts Eye and Ear. To date, 150 families have been enrolled in the study: 72 for surveys only (no genetic analyses), 16 for surveys and DHH genes only (no ACMG SF v3.0 analyses), and 62 for surveys (DHH genes and ACMG SF v3.0). Feedback from parents has been positive, with 61.5% acknowledging the benefits of receiving genome sequencing results on themselves, 67.6% acknowledging the benefits to both themselves and their child, and 62.6% of parents acknowledging that the genetic sequencing results have helped them understand the cause(s) of their child’s DHH diagnosis (Figure 1). These data demonstrate that the provision of a genetic diagnosis for DHH provides peace of mind to parents, even though personalised treatments are not currently available. Parents appreciate the complexity of receiving a genetic diagnosis and welcome the long-term investment to the potential new therapies for deafness.
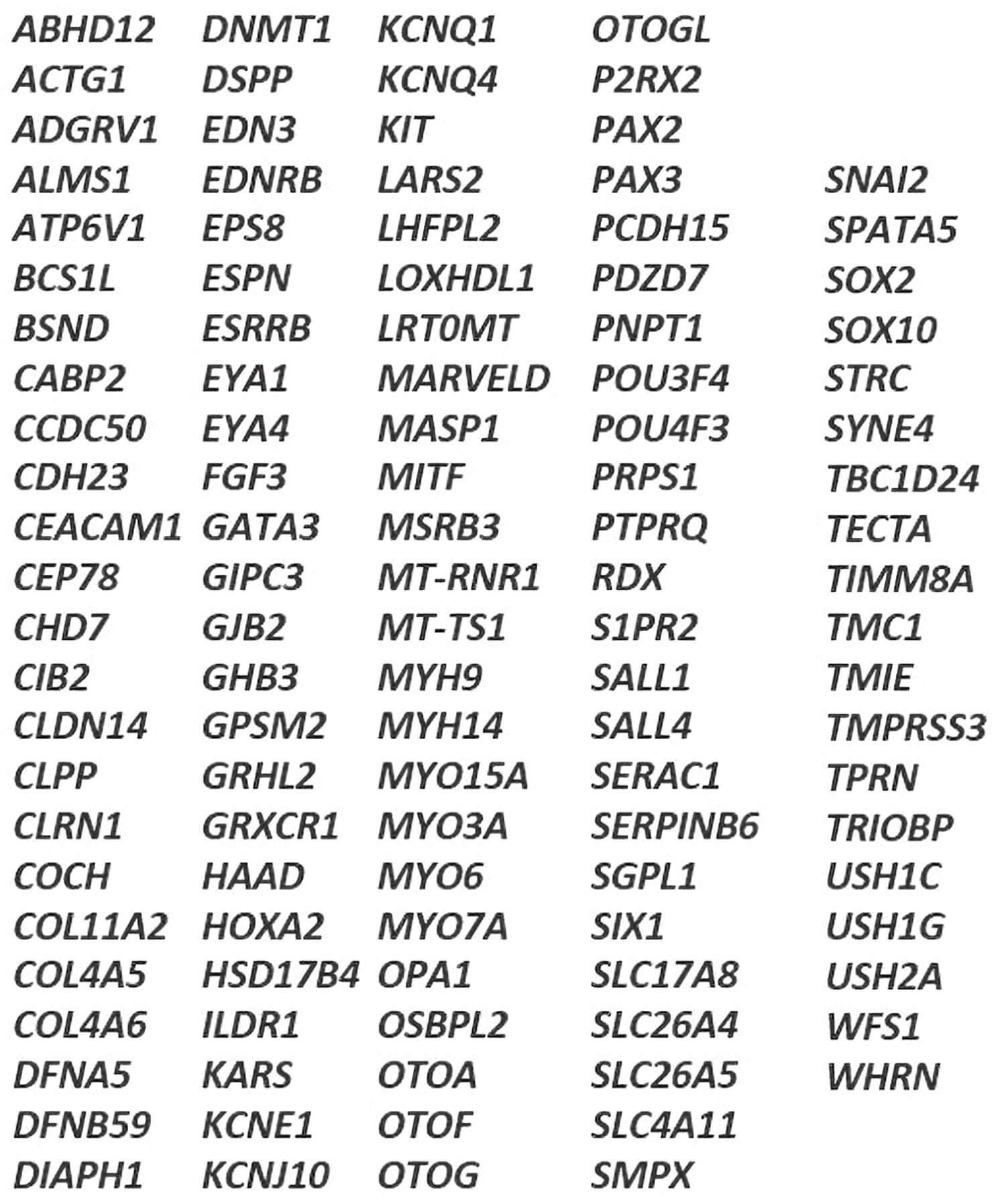
Figure 2. The UK National Health Service (NHS) non-syndromic hearing loss gene panel.
The 115 gene non-syndromic hearing loss panel will be offered to parents
whose baby is identified as deaf or hard of hearing.
SEQaBOO Manchester
SEQaBOO Manchester uses gene panel screening through the non-syndromic hearing loss panel (115 DHH genes, Figure 2) recently introduced into the UK’s National Health Service (NHS). SEQaBOO Manchester has only recently received approval to start enrolment and is recruiting from four Manchester University NHS Trust hospitals: Manchester Royal Infirmary, Trafford General Hospital, Altrincham Hospital and Withington Community Hospital. Participants who are approached at their confirmatory audiology appointment may enrol for surveys only - if the baby does not have a hearing loss - or for surveys and the gene panel, where the baby is identified as DHH (Figure 3).
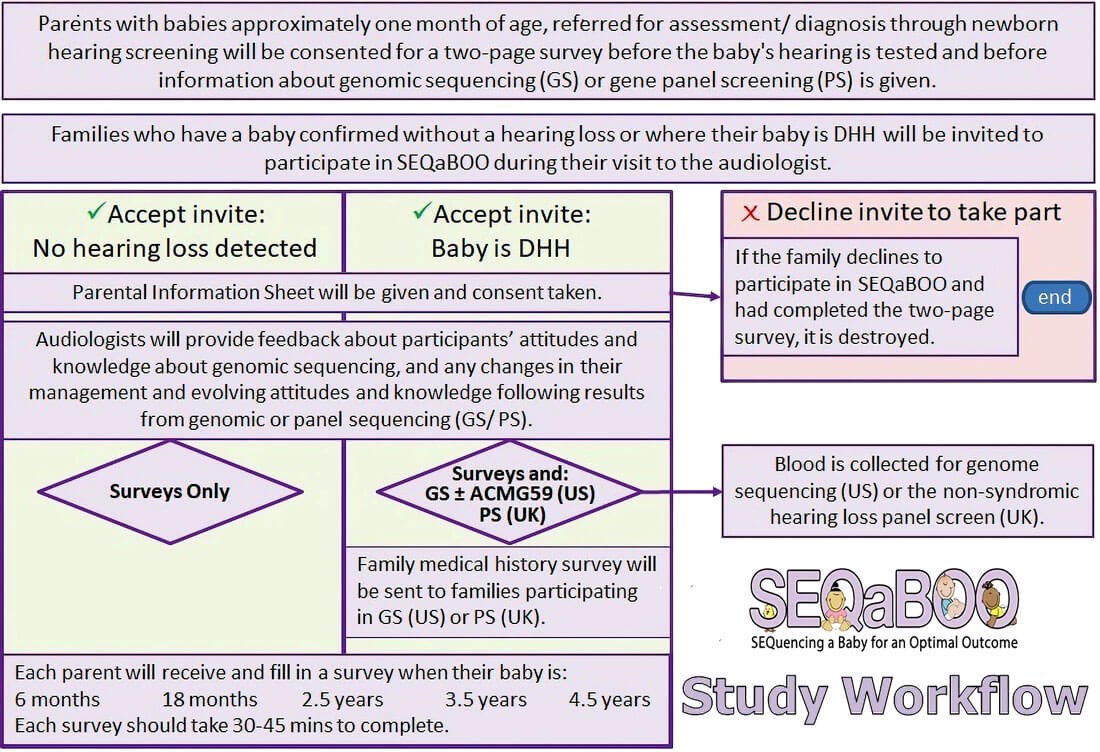
Figure 3. SEQaBOO enrolment pathway at confirmatory audiology testing.
SEQaBOO annual surveys and wider aims
Further to providing a genetic diagnosis for a baby’s deafness, SEQaBOO will explore the impact of that genetic diagnosis on parents and family members from clinical and social perspectives, through annual surveys, collected when the baby is six months of age and then each year, for four years. The surveys cover general health and the status of hearing, speech and language development. Parental knowledge and opinions of genomic testing are also considered to help us understand the best way of bringing genomic testing into newborn screening for the wider public.
“These data demonstrate that the provision of a genetic diagnosis for DHH provides peace of mind to parents, even though personalised treatments are not currently available”
Alongside SEQaBOO genomic sequencing, new bioinformatics tools and a user interface are being developed to provide a platform that can be easily used by clinical teams to re-evaluate data from newborns and infants with inconclusive or ambiguous findings. It will determine the significance of genomic screening through assessment of medical records, clinical testing, management strategies, and therapeutic interventions, and will inform development of a hearing health genetic registry [6] that will empower patients to become proactively involved in their hearing healthcare pathway.
The need for a hearing health genetic registry
Registries such as the Cystic Fibrosis registry have shown the value of including patients’ genetic information in the design and development of personalised treatments. They with Registries can provide a way for patients to be involved with their own healthcare pathway through a safe space for patient, healthcare professional, biotechnology and pharmaceutical company interactions. The large amount of variation in the DHH genetic landscape, as captured on the Deafness Variation Database (https://deafnessvariationdatabase.org), demands such an interactive environment to enable the partnering of genetically stratified DHH patients with novel therapeutics for inner ear and central hearing disorders [7]. Currently, there is no way for patients to be informed about developing treatments or therapies; conversely, innovative biotechnology/biopharmaceutical companies cannot access patients with specified genotypes for clinical trials.
Summary
Genome-based treatments are showing significant promise in medical health, and the development of precise, targeted cell- and gene-based therapies to restore or improve hearing will transform the lives of patients who are DHH. To this end, the genetic diagnoses that SEQaBOO provides to newborns and their families is world-leading in the transformation of providing personalised treatment pathways for patients who are DHH and will modernise the current approach with hearing aids and cochlear implants.
References
1. Shearer AE, Shen J, Amr S, et al. Newborn Hearing Screening Working Group of the National Coordinating Center for the Regional Genetics Networks. A proposal for comprehensive newborn hearing screening to improve identification of deaf and hard-of-hearing children. Genet Med 2019;21(11):2614-30.
2. Shearer AE, Hansen MR. Auditory synaptopathy, auditory neuropathy, and cochlear implantation. Laryngoscope Investig Otolaryngol 2019;4(4):429-40.
3. Stark Z, Dolman L, Manolio TA, et al. Integrating Genomics into Healthcare: A Global Responsibility. The American Journal of Human Genetics 2019;104:13-20.
4. Sloan-Heggen CM, Bierer AO, Shearer AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 2016;135(4):441‑50.
5. Miller DT, Lee K, Chung WK, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021 [Online ahead of print].
6. Short AD, Reghunathan SM, Morton CC. HEAR’s Why We Need a Hearing Health Genetic Registry in England. 2021. Submitted.
7. Schilder AGM, Su MP, Blackshaw H, et al. Hearing Protection, Restoration, and Regeneration: An Overview of Emerging Therapeutics for Inner Ear and Central Hearing Disorders. Otol Neurotol 2019;40(5):559-70.
Acknowledgements: ADS and CCM acknowledge support by the NIHR Manchester Biomedical Research Centre. CCM is also supported by the National Institutes of Deafness and Other Communication Disorders, National Institutes of Health (DC015052).